A good table can be visualised quickly
The more I work on data visualization using R, the more I realized that a good table has a clean structure and requires much less effort to be plotted out.
In ggplot, this often means that if a table requires more than a melt() in {reshape2} package then the table is not good enough - needs some more cleaning.
Working example:
d1 <- iris
d1 %>% head()
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
d1_melted <- d1 %>% melt(id.vars = c("Species"))
d1_melted %>% head()
## Species variable value
## 1 setosa Sepal.Length 5.1
## 2 setosa Sepal.Length 4.9
## 3 setosa Sepal.Length 4.7
## 4 setosa Sepal.Length 4.6
## 5 setosa Sepal.Length 5.0
## 6 setosa Sepal.Length 5.4
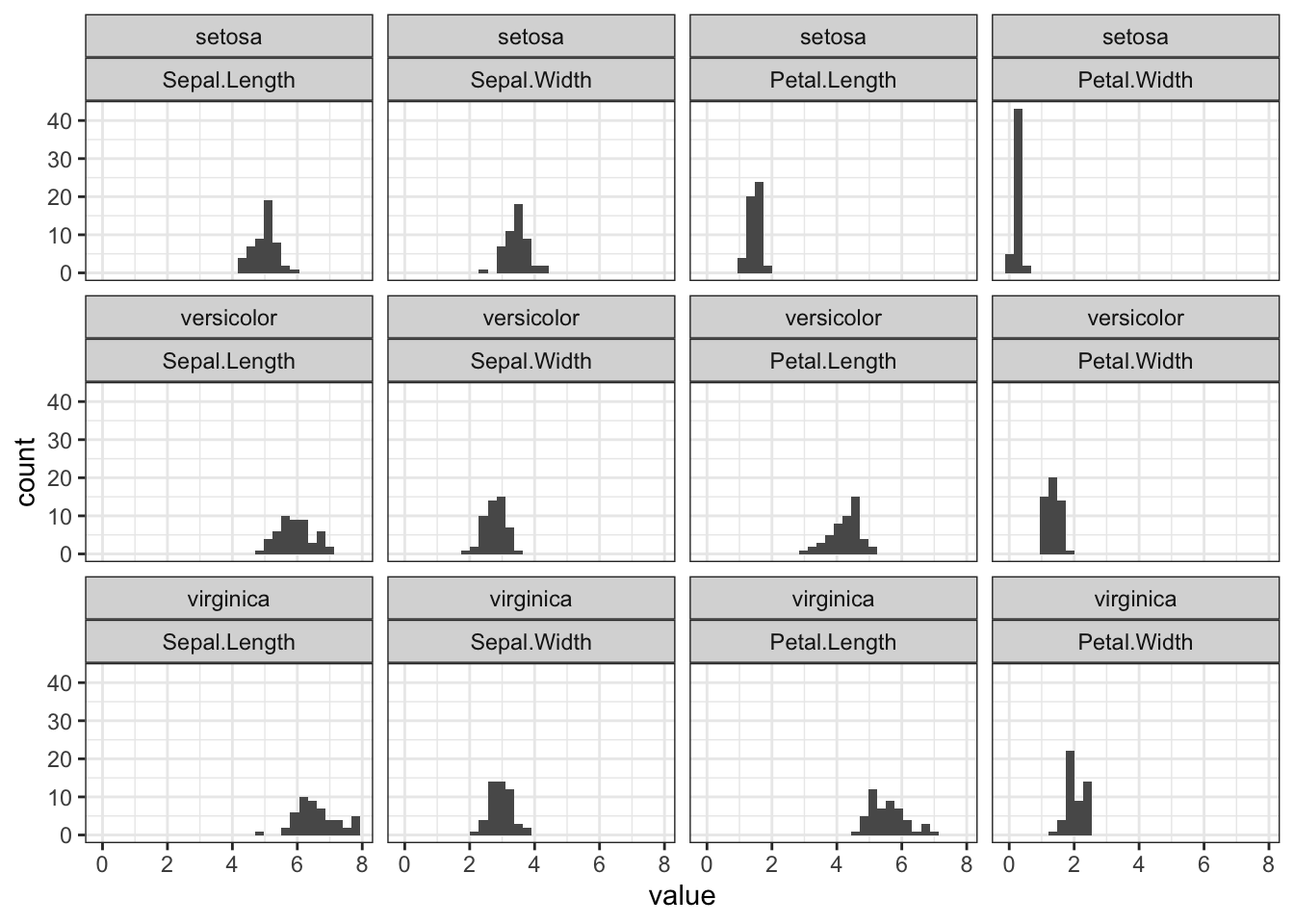
The facet_wrap() function can create subplots according to defined attributes. Here, I’d like to:
- display different
Speciesvertically, and - along each row to plot out length and with of sepal and petal. Now that these data are stored in
variableandvaluefrom themelt(), they can be specified in ggplot expression.
d1_melted %>% ggplot(aes(x=value)) + geom_histogram() +
facet_wrap(Species ~ variable) +
theme_bw()
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
Take home message: Be goal-oriented and thinking backward. Better-engineered data allows visualization and decision-makingm more efficiently.